
Research Highlights
Check out some of the recent discoveries from our labs
Weeraratna Lab Research

Age-dependent loss of HAPLN1 erodes vascular integrity via indirect upregulation of endothelial ICAM1 in melanoma. Marino-Bravante GE, et al. Nature Aging, 2024.
The Weeraratna lab studies melanoma and the impacts of aging. New research led by former BMB PhD student Gloria Marino Bravante shows that the secreted extracellular matrix protein HAPLN1, which is important in slowing melanoma progression but lost during aging, has an indirect role in maintaining blood vessel barrier function in melanoma tumors. These findings highlight the need to consider age and other variables when designing pre-clinical studies.
Prior studies showed that melanoma tumors are more vascularized in aged than young mice. When the team gave aged mice recombinant HAPLN1, the tumor vascularity levels were similar to young mice. They found the intratumor blood vessels in aged mice had lower levels of VE-cadherin, a protein involved in blood vessel barrier function; this was also reversed by recombinant HAPLN1. The team found that HAPLN1 loss leads to stiffer extracellular matrix, which increased expression of ICAM1, part of a signaling cascade that impacts VE-cadherin and vascular permeability. High vascularity and loss of barrier function are associated with metastasis, and the team found that aged mice given anti-ICAM1 therapy had smaller and less vascular primary melanoma tumors and fewer discernable lung metastases.
Find more in the Nature Aging paper published March 12, 2024.
Rebecca Lab Highlight

Jagirdar et al. ERK hyperactivation serves as a unified mechanism of escape in intrinsic and acquired CDK4/6 inhibitor resistance in acral lentiginous melanoma. Oncogene, 2023.
The Rebecca lab has identified how acral lentiginous melanoma gains resistance to CDK4/6 inhibitors and identified a potential way to counter it. ALM is a rare but particularly deadly form of melanoma with limited treatment options. About 60% of ALM patients have genetic alterations in the CDK4/6 pathway, but a recent clinical trial with a CDK4/6 inhibitor showed limited improvements in cancer progression and suggested that the cancer cells found ways to resist the drug.
Led by senior research scientist Kasturee Jagirdar, the researchers identified two resistance mechanisms that worked together: hyperactivation of the MAPK signaling pathway, which in turn drove upregulation of Cyclin D1, the co-activator of CDK4 and CDK6. When they paired the CDK4/6 inhibitor with a drug targeting the MAPK pathway, it was more effective than either drug alone, and in mouse models reduced tumor growth more than two-fold compared to single-agent treated mice. They also found that in ALM cells, CDK4/6 protein expression did not correlate well with predictions based on the CDK4/6 pathway genetic mutations. This suggest that to determine which ALM patients are most likely to benefit from CDK4/6 inhibitors, protein expression testing may be needed in addition to tumor sequencing.
Find full details in the December 8, 2023 Oncogene paper.
Kavran Lab Research
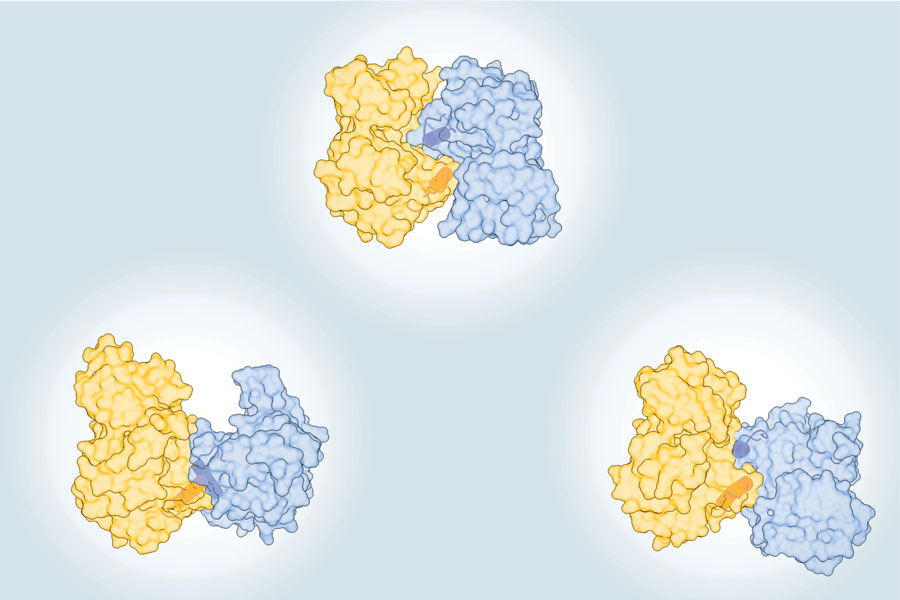
Dimerization and autophosphorylation of the MST family of kinases are controlled by the same set of residues. Weingartner, Tran, et al, Biochemical Journal, 2023.
In the Biochemical Journal's August 2023 cover story, Jennifer Kavran’s lab demonstrated that activation of the protein kinase MST1/2 by autophosphorylation requires dimerization, an activation mechanism that is likely conserved in similar kinases. These findings predict that activation of MST kinases could be blocked by regulatory proteins that prevent this dimerization.
MST1/2 kinases are a key part of the Hippo signaling pathway. They are activated by trans-autophosphorylation: one MST1/2 kinase must be phosphorylated by another one. Led by PhD student Kyler Weingartner and postdoc Thao Tran, the researchers resolve multiple different models of MST dimerization and autophosphorylation and identify the molecular mechanism of activation.
By analyzing the structures of a number of related MST family kinases they identified the aG-helix as the common feature involved in dimerization. To validate their model, they created mutant versions of MST2 with parts of the aG-helix replaced with alanines. These mutant kinases could phosphorylate the substrate MOB1A, but they did not dimerize and had very low levels of autophosphorylation.
Their analysis suggested this was a conserved feature, and they showed that aG-helix was required for autophosphorylation in another related kinase: Hippo.
For full details, read the Biochemical Journal paper, published August 16, 2023.
Culotta Lab Research

The role of manganese in morphogenesis and pathogenesis of the opportunistic fungal pathogen Candida albicans. Wildeman AS, Patel NK, Cormack BP, Culotta VC. PLoS Pathogens, 2023.
Valeria Culotta’s lab has uncovered a role for manganese in Candida albicans infections: not only do the fungal cells need the metal micronutrient for certain aspects of their cellular biology and pathogenesis, infected mice appear to reduce Mn levels in a key tissue.
The Culotta lab has long studied nutritional immunity, where hosts withhold certain micronutrients including metals during infection. Unlike Cu, Zn, and Fe, the role of Mn in fungal infections hasn’t been explored as much. Led by PhD student Asia Wildeman, the Culotta lab examined the role in C. albicans.
When they knocked out either of two Mn transporters, Smf12 and Smf13, intracellular levels of Mn, but not Cu, Zn, or Fe, dropped, and these mutants had defects in certain enzyme activities and hyphal filament formation.
The researchers examined pathogenesis using a mouse model where the kidneys were the main infection site. They found that the Mn levels in kidney tissues of infected mice dropped by up to 40%, the first report of whole tissue reduction of Mn as a host response to fungal infection. Further, mice infected with the Mn transporter mutants had a higher survival rate and less lost less weight than the controls.
Find more details in the PLoS Pathogens paper, published July 7, 2023.
Leung Lab Research

Switch-like compaction of poly(ADP-ribose) upon cation binding. Badiee et al, 2023 PNAS.
Anthony Leung's lab has teamed up again with Sua Myong's lab and discovered unexpected structural properties of poly(ADP-ribose). PAR is a nucleic acid-like molecule that regulates crucial cellular processes through protein modification, and can be a scaffold of molecular complexes. This study, led by postdoc Mohsen Badiee, revealed that PAR chains maintain rigidity until reaching a certain salt concentration, then abruptly compact, unlike DNA and RNA which gradually become more compact as the concentration increases.
They also looked at the response to FUS, a PAR binding protein with positively charged domains. Previously they discovered that PAR is a potent trigger of FUS condensation. Here they showed that increasing FUS concentrations triggered the switch from stiff to compact PAR. In contrast, a different PAR-binding protein did not alter PAR stiffness.
These findings imply that a given PAR molecule can adopt various shapes based on environmental factors and provides insights into how proteins interact with PAR. Expanding our understanding of PAR’s structural behavior is important to untangling its role in normal cellular processes and in diseases like cancer and neurodegenerative diseases.
For more details, read the full paper at PNAS, or learn about recent advances in PAR research tools in the lab’s recent Molecular Cell review.
Wang Lab Research

DNA-initiated epigenetic cascades driven by C9orf72 hexanucleotide repeat, Liu et al, 2023, Neuron
Jiou Wang’s lab has shown that a repeat expansion mutation linked to neurodegeneration may cause disease through its effects triggered directly by the DNA, independent of the mutation’s downstream effects on RNA and protein that have been demonstrated by his lab and others.
The C9orf72 gene normally has two to ten “GGGGCC” repeats in a non-coding region, but the hexanucleotide repeat expansion mutations have hundreds to thousands. Led by postdoctoral fellow Yang Liu, PhD, the lab showed that a DNA-binding protein called DAXX preferentially binds to this much longer repeat region and accumulates in the cell.
The accumulation of DAXX has a cascade of pathologic effects: it triggers a liquid-liquid phase separation that drives chromatin structure and epigenetic modifications changes, suppresses the normal stress-dependent induction of C9orf72, and influences global gene expression. These findings may open up new prevention or treatment strategies for neurodegenerative diseases, and in fact the researchers found that reducing DAXX or rebalancing the epigenetic modifications reduced the sensitivity of patient neurons to stress.
For more details about the research, check out the paper at Neuron.
Matunis Lab Research

Analysis of a degron-containing reporter protein GFP-CL1 reveals a role for SUMO1 in cytosolic protein quality control, Wang et al, 2023, Journal of Biological Chemistry
Mike Matunis' lab has uncovered a new role for small ubiquitin-related modifiers, or SUMO, in protein quality control, the surveillance mechanisms that cells use to identify and degrade misfolded proteins – for the first time showing a role in PQC in the cytoplasm, rather than nucleus. The accumulation of misfolded proteins can cause proteotoxic stress that can damage cells and is associated with multiple human diseases, including neurodegeneration, and these findings may provide insights into the role of sumolyation in those diseases.
Led by BMB PhD student Wei Wang, the researchers showed that in the yeast Saccharomyces cerevisiae and in humans, a SUMO protein is required for the timely turnover of a PQC reporter protein containing the CL1 degron, a short degradation signal that can misfold and trigger rapid degradation of the protein. SUMO only affected the turnover in the cytoplasm, not the nucleus. Experiments examining different aspects of the CL1 degron response pathway showed that SUMO’s role is upstream of the ubiquitylation and proteasome activity steps, and suggest that it promotes degradation by maintaining the solubility of the reporter proteins, but more research is needed to fully understand the mechanisms involved.
For more details check out the paper at the Journal of Biological Chemistry.
Wang Lab Research

Intracellular energy controls dynamics of stress-induced ribonucleoprotein granules, Wang et al, 2022, Nature Communications
Jiou Wang’s lab studies fundamental mechanisms of neurodegenerative diseases, which are often associated with cellular stress and disrupted energy metabolism. They have discovered a unique type of ribonucleoprotein granule that can be formed under energy deficiency stress conditions.
Stress granules, or SGs, comprising condensed proteins and RNAs, are a type of membraneless organelle induced by certain stressors. Researchers studying cells with disrupted energy metabolism found a new type of stress-induced granule: energy deficiency-induced stress granules. These eSGs have a distinct protein composition and higher RNA contents, suggestive of different functions. They also assemble differently, independent of translation factor eIF2α phosphorylation that is a hallmark of conventional SGs.
Neurons derived from patients with a major form of motor neuron degenerative disease, which have pre-existing metabolic disruptions, had abnormal eSG formation, suggesting its relevance to the disease processes. Full details, including more about eSG features and assembly, are in Intracellular energy controls dynamics of stress-induced ribonucleoprotein granules at Nature Communications.
Kavran Lab Research

Kinetic Regulation of the Mammalian Sterile 20-like Kinase 2 (MST2), Koehler, Tran, et al, 2022, Biochemistry.
Jennifer Kavran’s lab studies HIPPO: not the fierce but adorable semi-aquatic mammal, but a signaling pathway with roles that include tumor suppression.
Led by PhD student TJ Koehler and then-postdoc Thao Tran, the lab studied kinetic regulation of key Hippo kinase MST2. Examining the activity of different versions of MST2 under different conditions, they showed that by far the largest driver of MST2 activity was if it was phosphorylated, regardless of which domains were present.
They revealed a novel role of the C-terminal SARAH domain: it dramatically increased activity in unphosphorylated proteins, but reduced it in phosphorylated proteins, with evidence suggesting that it disrupted interactions with ATP. They also found that some factors that affect MST 1/2 activity in cells, such as caspase cleavage, had little impact on this in vitro activity, suggesting that these factors influence non-kinetic aspects of the pathway in vivo, such as cellular localization.
Check out the paper in Biochemistry for all the details!
